TI Opti Taq DNA Polymerase
TI-inhibited "HotStart" Taq + Pyrococcus sp. DNA Polymerase for high yield, robust PCR amplification.
- Robust, difficult & long PCR.
- Moderate "proofreading".
- Separate enzyme and 10x buffers.
- Contains both, non-colored and colo
Detailed Product Description
English Version
Deutsche Version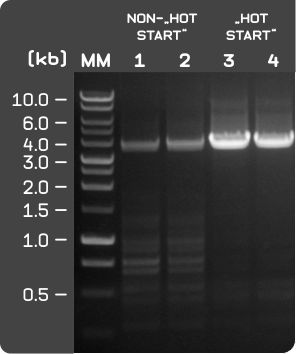
Figure 1: PCR amplification using EURx TI OptiTaq DNA Polymerase - Forced "HotStart" demonstration.
A 4 kb amplicon of the human alpha-globin gene was amplified using EURx non-"Hot Start" OptiTaq DNA Polymerase and "Hot Start" tiOptiTaq DNA Polymerase, both in 10 x Pol Buffer B and 0.2 mM dNTPs within a 50 µl reaction volume.
For demonstrating "HotStart" capabilities, best conditions for (normally undesired) primer dimer formation were forced by incubating assembled reactions for 30 min at 25°C prior to start of PCR cycling - admittedly somewhat exaggerated conditions, but this is supposed to be an "extreme case" demonstation.
At ambient temperature, residual activity of Taq DNA polymerase would, if not blocked, lead to ample formation of small dsDNA molecules infamously known as primer dimers. It has to be kept in mind, that Taq activity at ambient temperature correlates to as much as ∼10% of Taq activity at 72°C.
Under such conditions, non-specific primer annealing would extend randomly annealing PCR primers to double stranded DNA molecules, thus generating many, many short double stranded DNA fragments with perfectly matching priming sites.
These small DNA molecules would efficiently amplify during PCR, thus competing with larger DNA amplicons for PCR resources and finally leading to reduced PCR yield of the actually desired amplicon. Furthermore, co-amplification of a huge fraction of small, unwanted dsDNA molecules would disturb any subsequent cloning and clone picking steps.
"HotStart" enzymes, such as the TI enzyme series, would strongly inhibit formation of non-desired primer dimers by blocking DNA polymerase at ambient temperature. TI OptiTaq efficiently prevents (1) primer dimer formation and (2) PCR artifact generation due to non-specific annealing within the short, but critical time span between PCR reaction assembly and the time point, where the PCR assay has reached a temperature of >90°C during the initial denaturation step.- Lane M: molecular size marker- Perfect Plus 1 kb DNA Ladder (E3131).
- Lane 1,2: PCR amplification reactions using 1.25 U non-"HotStart" OptiTaq DNA Polymerase (E2600). Note the diminished yield of the desired 4 kb amplicon and the presence of additional, small bands, indicative for primer dimer formation as well as for non-specific primer-template DNA annealing.
- Lane 3, 4: PCR amplification reactions using 1.25 U tiOptiTaq DNA Polymerase. All reactions were incubated for 30 min at 25°C before PCR.

