Endocyclase N15
Endocyclase N15 (Protelomerase N15) catalyzes the formation of covalent links between complementary 5'‑ and 3'‑ ends (clamps DNA strands together) and forms exonuclease-protected hairpin ends at specific double-stranded DNA recognition sites.
Detailed Product Description
English Version
Deutsche Version
Applications
Mode of Action
Figure 1: Circularization and covalent capping of linear DNA (e.g. PCR amplicons).
Figure 2: Conversion of covalently closed circular plasmid DNA into two separate linear plasmids.
Figure 3: Experimental strategy for conversion of covalently closed plasmid DNA (ccDNA) to linear minicircle plasmid DNA. This procedure allows to remove unwanted bacterial gene sequences, which, upon transfer in eukaryotic cells, may lead to transgene expression silencing. Removal of unwanted bacterial DNA results in an elevated level of sustained transgene expression.
Sketch of a typical bacterial plasmid containing genes of interest. The area marked with red color is flanked by two endocyclase recognition sites and does contain the target payload genes. The area marked in black contains typical plasmid bacterial backbone sequences such as origin of replication (ori) and antibiotic resistance genes.
- Generate covalently closed clamps between two complementary DNA strands on one or on both ends.
- Convert PCR products to linear plasmids with capped "hairpin" ends.
- Convert circular plasmid DNA to linear plasmid DNA.
- Generate linear miniplasmid DNA from circular plasmid DNA. This allows to minimize introduction of bacterial backbone sequences to eukaryotic cells.
- Protects PCR-amplified. processed DNA from exonuclease attack and digestion.
- Transfection of cell cultures with linear plasmids generated directly from PCR products.
- Generation of linear BACs (Bacterial Artificial Chromosomes).
Mode of Action
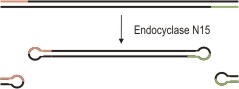
Figure 1: Circularization and covalent capping of linear DNA (e.g. PCR amplicons).
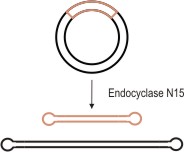
Figure 2: Conversion of covalently closed circular plasmid DNA into two separate linear plasmids.
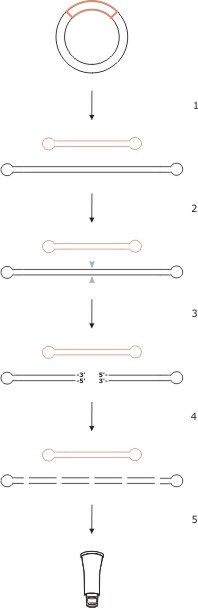
Figure 3: Experimental strategy for conversion of covalently closed plasmid DNA (ccDNA) to linear minicircle plasmid DNA. This procedure allows to remove unwanted bacterial gene sequences, which, upon transfer in eukaryotic cells, may lead to transgene expression silencing. Removal of unwanted bacterial DNA results in an elevated level of sustained transgene expression.
Sketch of a typical bacterial plasmid containing genes of interest. The area marked with red color is flanked by two endocyclase recognition sites and does contain the target payload genes. The area marked in black contains typical plasmid bacterial backbone sequences such as origin of replication (ori) and antibiotic resistance genes.
- (1) Processing of ccDNA with endocyclase yields two linear plasmids.
- (2) Digestion with a restriction enzyme. The restriction enzyme of choice cuts within the bacterial backbone plasmid only, but has no recognition site within the payload gene plasmid.
- (3) The bacterial backbone plasmid is opened and exposes free 5'- and 3'- ends of DNA at the restriction site.
- (4) A dsDNA-specific exonuclease attacks free DNA ends and digests all genes residing on the bacterial backbone plasmid to completeness. Since payload genes reside on a separate linear plasmid with covalently closed DNA ends, exonuclease cannot attack and cannot digest the plasmid containing the genes of interest.
- (5) The linear plasmid containing the genes of interest is purified and is ready to use.

